Plotting and Visualizing Simulated Output
Visualizing simulated results, is a critical step in understanding and communicating model behavior. Visualization serves as the blueprint of any experiment as it generates scientific insight underlying the data. The goal of this tutorial, therefore, is to help you quickly set up, diagnose, and report your simulation outcomes effectively, enabling clear interpretation and compelling presentation of your results.
from apsimNGpy.core.experimentmanager import ExperimentManager
from matplotlib import pyplot as plt
Create the experiment
model = ExperimentManager('Maize', out_path = 'my_experiment.apsimx')
# init the experiment
model.init_experiment(permutation =True)
Adding factors to the experiment
Adding factors to the experiment requires that we understand the model structure. This can be accomplished
by: inspect_model(), inspect_file(),
inspect_model_parameters_by_path()
Inspect the whole simulation tree
model.inspect_file()
└── Simulations: .Simulations
├── DataStore: .Simulations.DataStore
└── Experiment: .Simulations.Experiment
├── Factors: .Simulations.Experiment.Factors
│ └── Permutation: .Simulations.Experiment.Factors.Permutation
└── Simulation: .Simulations.Experiment.Simulation
├── Clock: .Simulations.Experiment.Simulation.Clock
├── Field: .Simulations.Experiment.Simulation.Field
│ ├── Fertilise at sowing: .Simulations.Experiment.Simulation.Field.Fertilise at sowing
│ ├── Fertiliser: .Simulations.Experiment.Simulation.Field.Fertiliser
│ ├── Harvest: .Simulations.Experiment.Simulation.Field.Harvest
│ ├── Maize: .Simulations.Experiment.Simulation.Field.Maize
│ ├── Report: .Simulations.Experiment.Simulation.Field.Report
│ ├── Soil: .Simulations.Experiment.Simulation.Field.Soil
│ │ ├── Chemical: .Simulations.Experiment.Simulation.Field.Soil.Chemical
│ │ ├── NH4: .Simulations.Experiment.Simulation.Field.Soil.NH4
│ │ ├── NO3: .Simulations.Experiment.Simulation.Field.Soil.NO3
│ │ ├── Organic: .Simulations.Experiment.Simulation.Field.Soil.Organic
│ │ ├── Physical: .Simulations.Experiment.Simulation.Field.Soil.Physical
│ │ │ └── MaizeSoil: .Simulations.Experiment.Simulation.Field.Soil.Physical.MaizeSoil
│ │ ├── Urea: .Simulations.Experiment.Simulation.Field.Soil.Urea
│ │ └── Water: .Simulations.Experiment.Simulation.Field.Soil.Water
│ ├── Sow using a variable rule: .Simulations.Experiment.Simulation.Field.Sow using a variable rule
│ └── SurfaceOrganicMatter: .Simulations.Experiment.Simulation.Field.SurfaceOrganicMatter
├── Graph: .Simulations.Experiment.Simulation.Graph
│ └── Series: .Simulations.Experiment.Simulation.Graph.Series
├── MicroClimate: .Simulations.Experiment.Simulation.MicroClimate
├── SoilArbitrator: .Simulations.Experiment.Simulation.SoilArbitrator
├── Summary: .Simulations.Experiment.Simulation.Summary
└── Weather: .Simulations.Experiment.Simulation.Weather
Inspect the full paths to the manager scripts in the model
model.inspect_model('Models.Manager')
['.Simulations.Experiment.Simulation.Field.Sow using a variable rule',
'.Simulations.Experiment.Simulation.Field.Fertilise at sowing',
'.Simulations.Experiment.Simulation.Field.Harvest']
Inspect the names to the manager scripts in the model
model.inspect_model('Models.Manager', fullpath=False)
['Sow using a variable rule', 'Fertilise at sowing', 'Harvest']
If we want to edit any of the above script in the experiment, there is need to inspect the associated underlying parameters
‘.Simulations.Experiment.Simulation.Field.Sow using a variable rule’
model.inspect_model_parameters_by_path('.Simulations.Experiment.Simulation.Field.Sow using a variable rule')
{'Crop': 'Maize',
'StartDate': '1-nov',
'EndDate': '10-jan',
'MinESW': '100.0',
'MinRain': '25.0',
'RainDays': '7',
'CultivarName': 'Dekalb_XL82',
'SowingDepth': '30.0',
'RowSpacing': '750.0',
'Population': '6.0'}
‘.Simulations.Experiment.Simulation.Field.Fertilise at sowing’
model.inspect_model_parameters_by_path('.Simulations.Experiment.Simulation.Field.Fertilise at sowing')
{'Crop': 'Maize', 'FertiliserType': 'NO3N', 'Amount': '160.0'}
Add factors along the the paramters; Amount and Population from the last and first scripts, respectively.
# Population
model.add_factor(specification='[Sow using a variable rule].Script.Population = 4, 6, 8, 10', factor_name='Population')
# Nitrogen fertilizers
model.add_factor(specification='[Fertilise at sowing].Script.Amount= 0, 100,150, 200, 250', factor_name= 'Nitrogen')
Before we run,let’s add one report variable for an easy demonstration
model.add_report_variable(variable_spec=['[Clock].Today.Year as year'], report_name='Report')
# run the model to generate output
model.run()
Inspect the simulated results
model.results.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 200 entries, 0 to 199
Data columns (total 19 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 CheckpointID 200 non-null int64
1 SimulationID 200 non-null int64
2 Experiment 200 non-null object
3 Population 200 non-null object
4 Nitrogen 200 non-null object
5 Zone 200 non-null object
6 Clock.Today 200 non-null object
7 Maize.Phenology.CurrentStageName 200 non-null object
8 Maize.AboveGround.Wt 200 non-null float64
9 Maize.AboveGround.N 200 non-null float64
10 Yield 200 non-null float64
11 Maize.Grain.Wt 200 non-null float64
12 Maize.Grain.Size 200 non-null float64
13 Maize.Grain.NumberFunction 200 non-null float64
14 Maize.Grain.Total.Wt 200 non-null float64
15 Maize.Grain.N 200 non-null float64
16 Maize.Total.Wt 200 non-null float64
17 year 200 non-null int64
18 source_table 200 non-null object
dtypes: float64(9), int64(3), object(7)
memory usage: 29.8+ KB
# By default, the parameter name of each factor is also populated in the data frame.
Statistical results for each column
model.results.describe()
CheckpointID SimulationID Maize.AboveGround.Wt Maize.AboveGround.N \
count 200.0 200.000000 200.000000 200.000000
mean 1.0 10.500000 1067.600206 10.239469
std 0.0 5.780751 607.686584 6.299485
min 1.0 1.000000 51.423456 0.320986
25% 1.0 5.750000 708.023479 5.095767
50% 1.0 10.500000 987.749088 9.423852
75% 1.0 15.250000 1579.116949 15.402366
max 1.0 20.000000 2277.481374 22.475693
Yield Maize.Grain.Wt Maize.Grain.Size \
count 200.000000 200.000000 200.000000
mean 4765.995363 476.599536 0.245769
std 3093.295392 309.329539 0.073753
min 0.000000 0.000000 0.000000
25% 2535.993006 253.599301 0.223099
50% 3894.863108 389.486311 0.274101
75% 7819.712885 781.971288 0.299073
max 10881.111792 1088.111179 0.315281
Maize.Grain.NumberFunction Maize.Grain.Total.Wt Maize.Grain.N \
count 200.000000 200.000000 200.000000
mean 1890.470249 476.599536 6.187685
std 1077.127429 309.329539 4.118276
min 0.000000 0.000000 0.000000
25% 979.352980 253.599301 3.051965
50% 1855.994293 389.486311 5.112196
75% 2861.250716 781.971288 10.372698
max 3726.740206 1088.111179 13.852686
Maize.Total.Wt year
count 200.000000 200.000000
mean 1167.632796 1995.500000
std 656.486381 2.879489
min 53.721510 1991.000000
25% 774.862931 1993.000000
50% 1104.093752 1995.500000
75% 1705.820275 1998.000000
max 2457.083319 2000.000000
Moving average plots
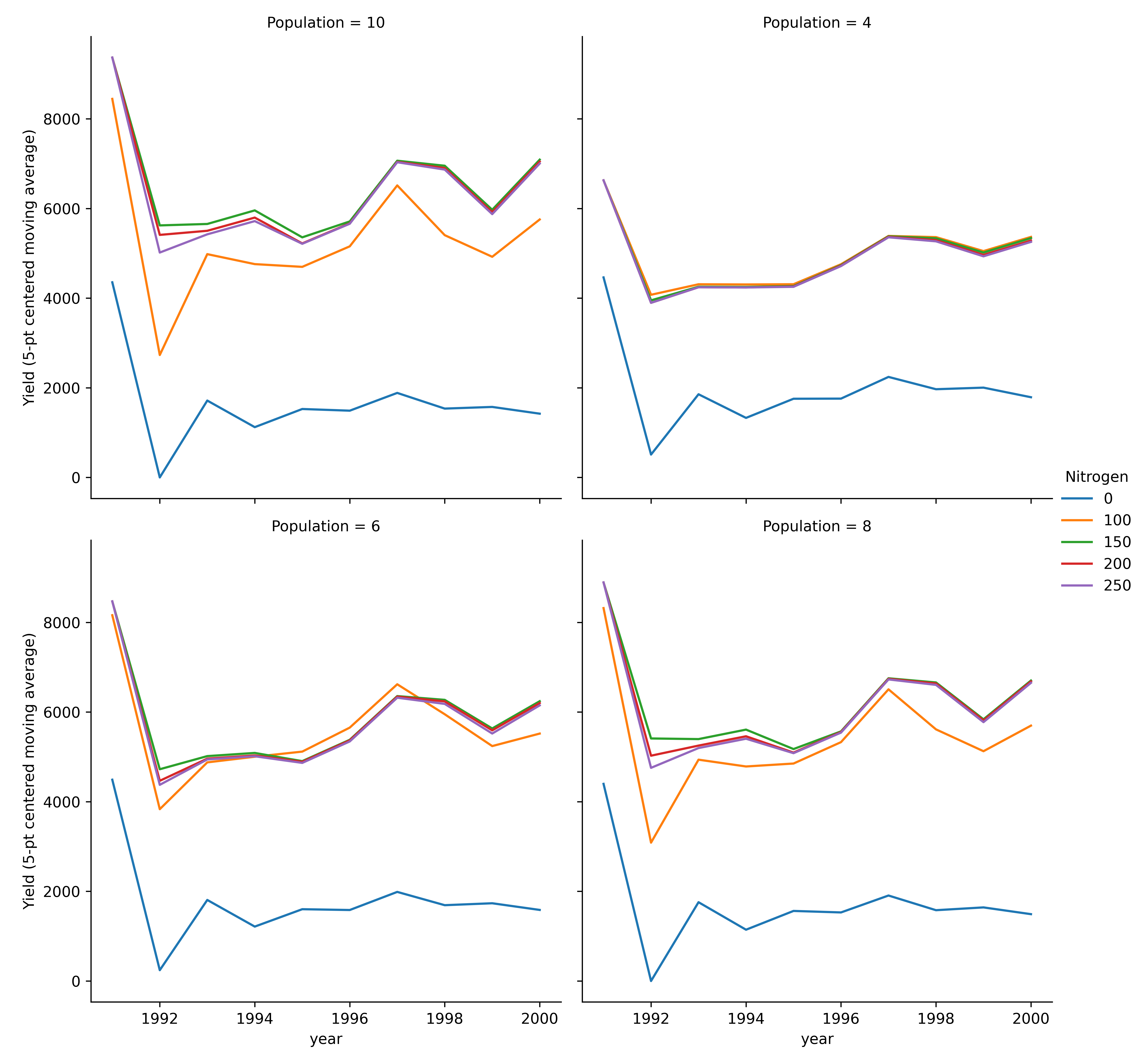
model.plot_mva(table='Report', response='Yield', time_col='year', col_wrap=2, palette='tab10',
errorbar=None, estimator='mean', grouping=('Amount', 'Population'), hue='Nitrogen', col='Population')
Multi-year moving average for each experiment (line plot).
Categorical Plots
Box plots
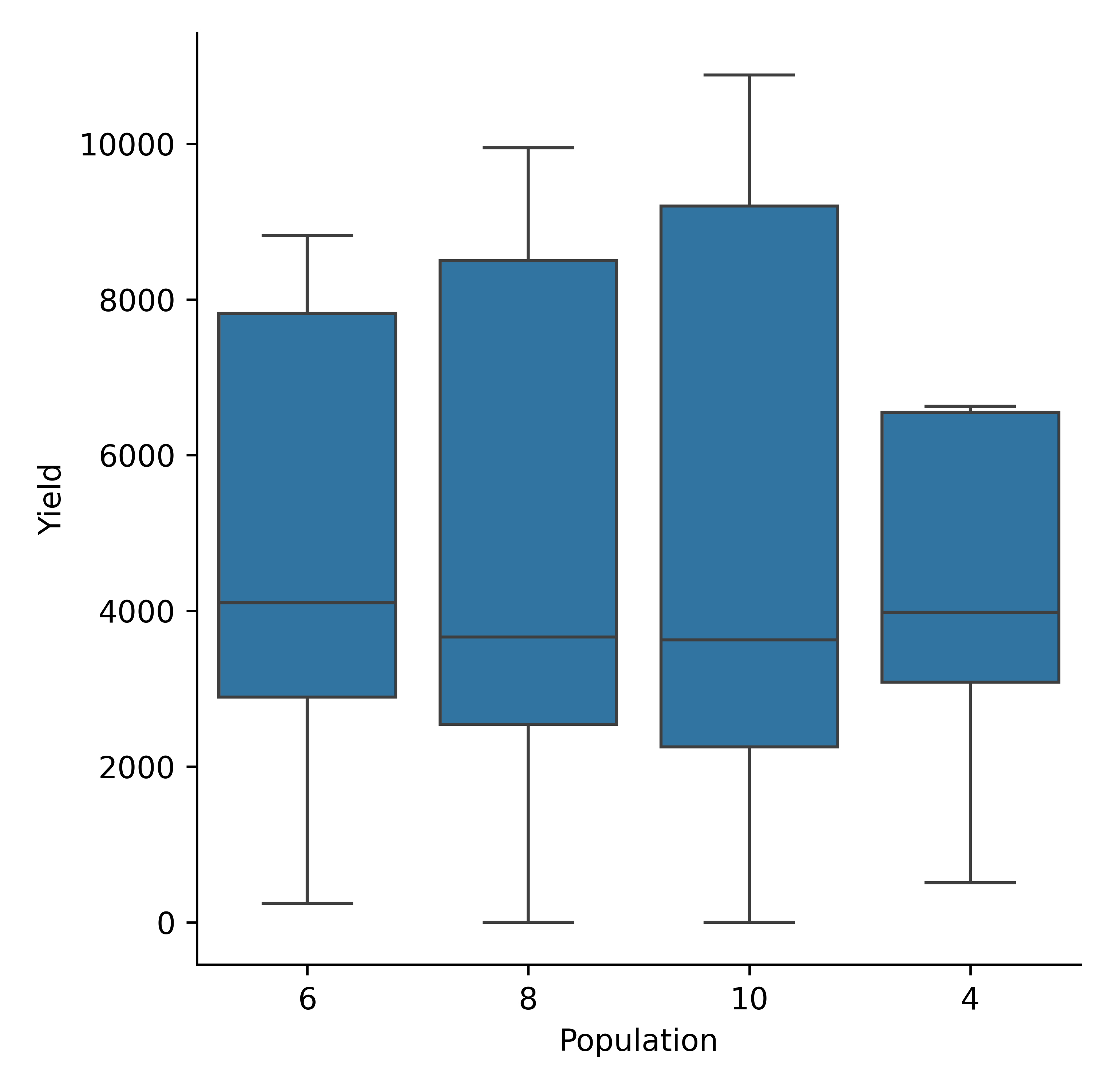
model.cat_plot(table = 'Report', y='Yield', x= 'Population', kind = 'box')
Maize yield variability by population density (mva plot).
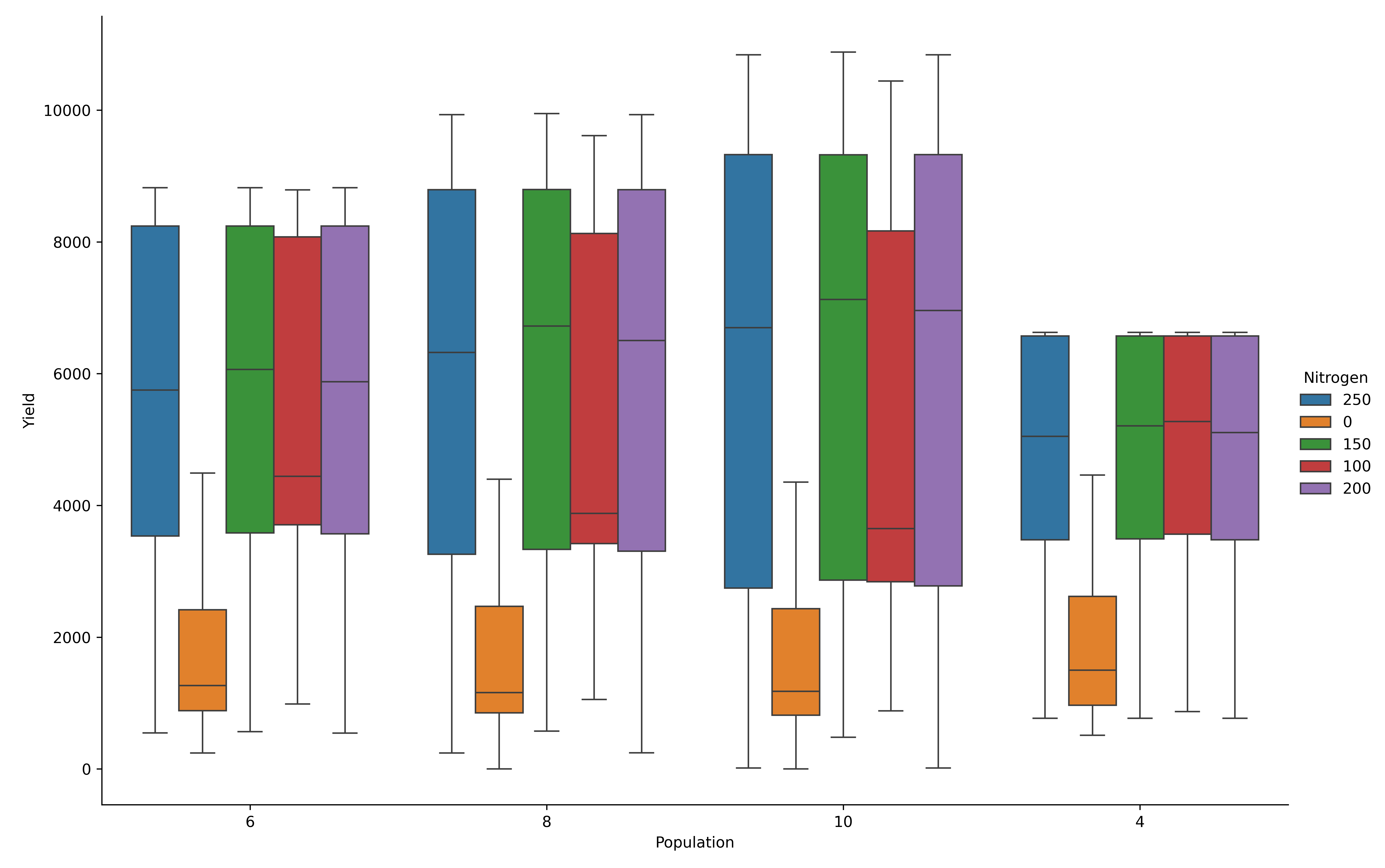
Add Nitrogen fertilizer as hue for contrast across the nitrogen treatments
model.cat_plot(table = 'Report', y='Yield', x= 'Population', palette='tab10',
kind = 'box', hue= 'Nitrogen', height=8, aspect=1.5)
plt.savefig(dir_p/’hue_nitrogen.png’, dpi=600)
Maize yield variability by population density and nitrogen fertilizer (box plot).
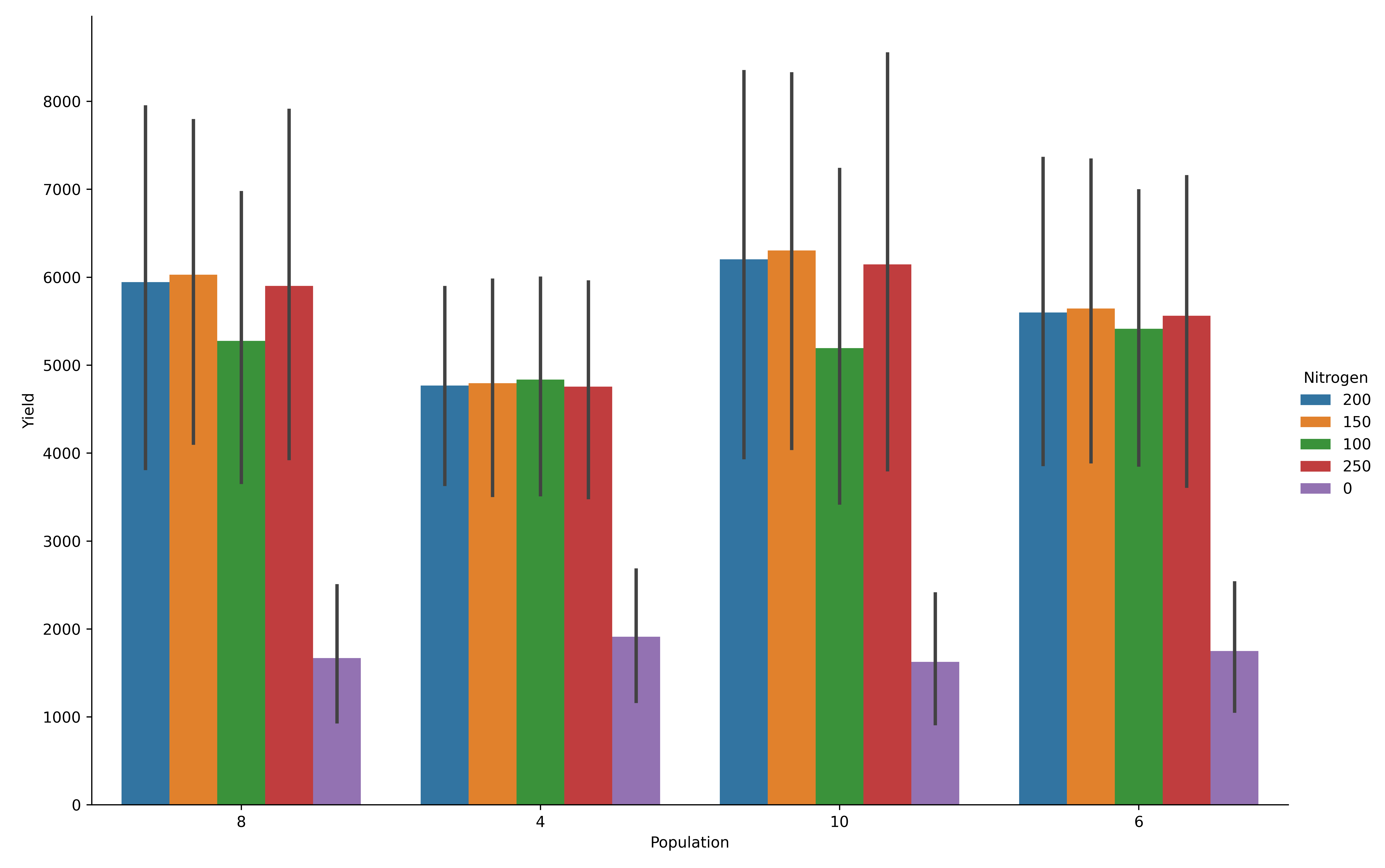
Bar Plots
model.cat_plot(table = 'Report', y='Yield',
x= 'Population', kind = 'bar', hue= 'Nitrogen', height=8, aspect=1.5)
Maize yield variability by population density (bar plot).
Changing statistical estimators. The example below shows how to switch estimators, and after the change to sum, the y-axis is now inflated
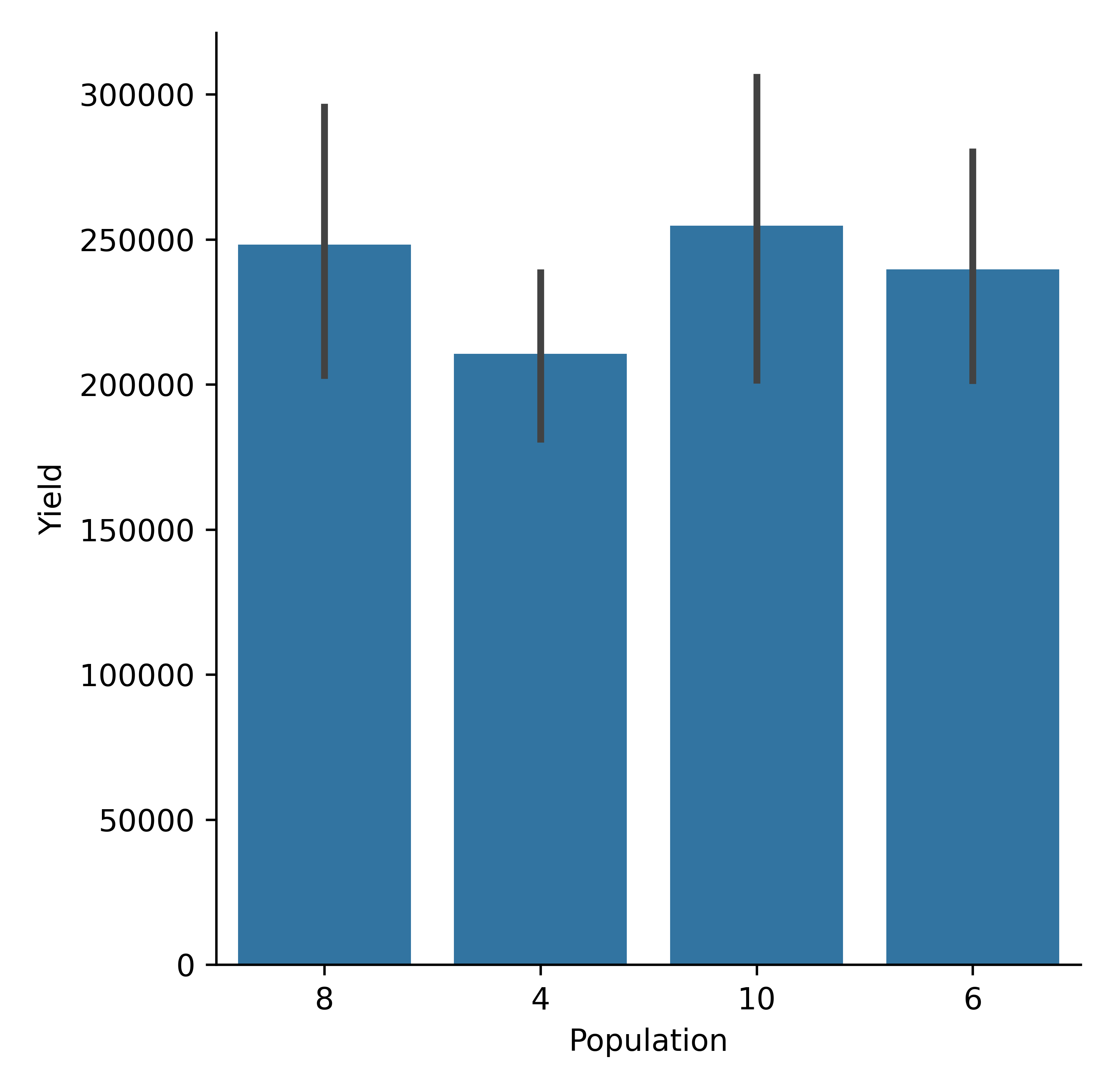
model.cat_plot(table = 'Report', y='Yield', x= 'Population', kind = 'bar', estimator='sum')
Maize yield variability by population density (bar plot, estimator =sum).
If you want to plot in a different unit or apply on-the-fly calculations, all plotting methods allow simple mathematical expressions to be passed, as shown below.
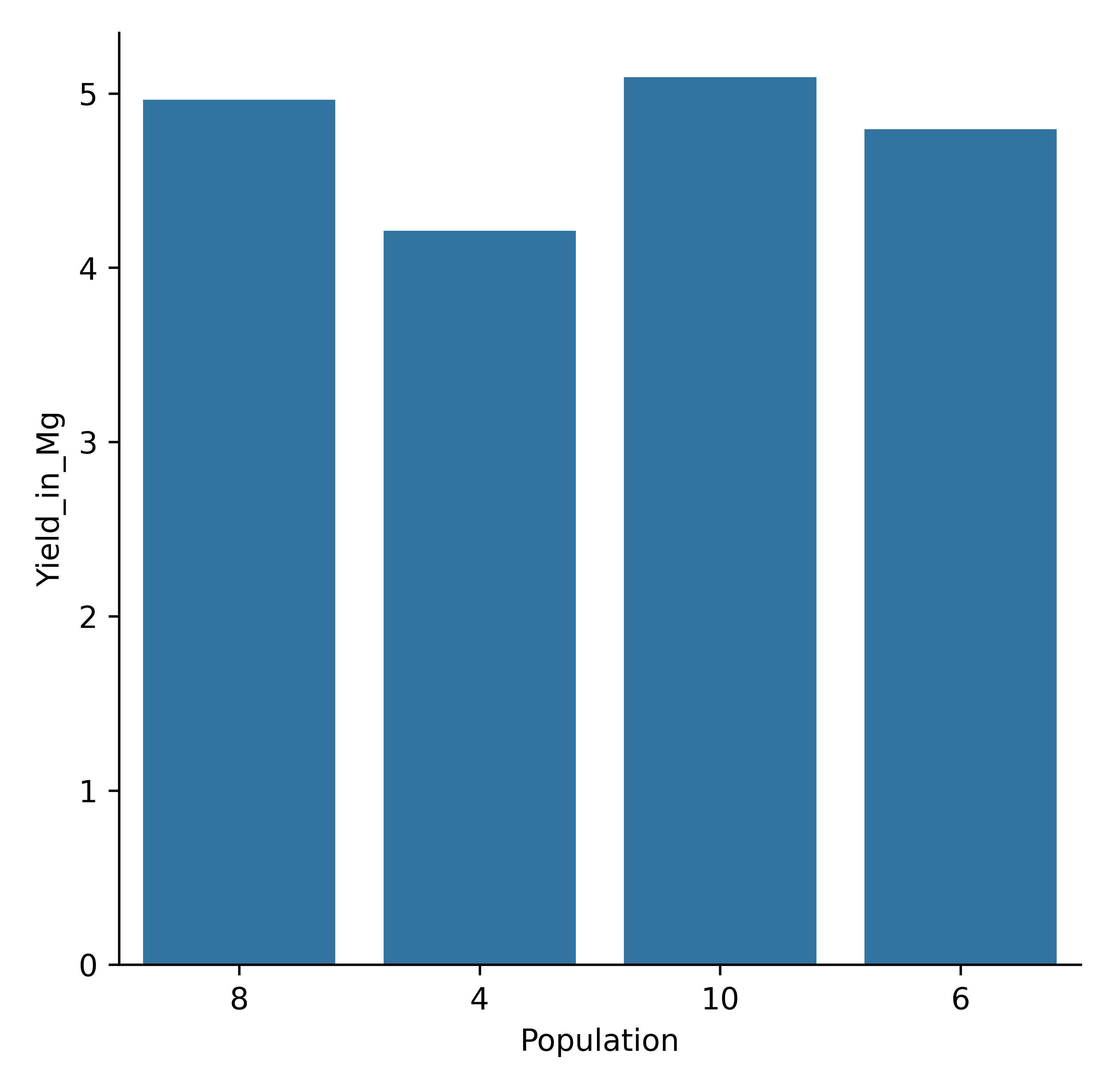
model.cat_plot('Report', expression="Yield_in_Mg = Yield/1000",
y='Yield_in_Mg', x= 'Population', kind = 'bar', errorbar=None)
Maize yield variability by population density in Mg (bar plot, estimator =sum).
Tidy up the plots for reporting
The heavy lifting is done; now polish the figure—adjust labels, titles, and size. All plotting methods on ApsimModel and ExperimentManager return a seaborn.axisgrid.FacetGrid, so you can keep customizing afterward. Because Seaborn sits on Matplotlib, any Matplotlib styling you pass (or apply later) still works.
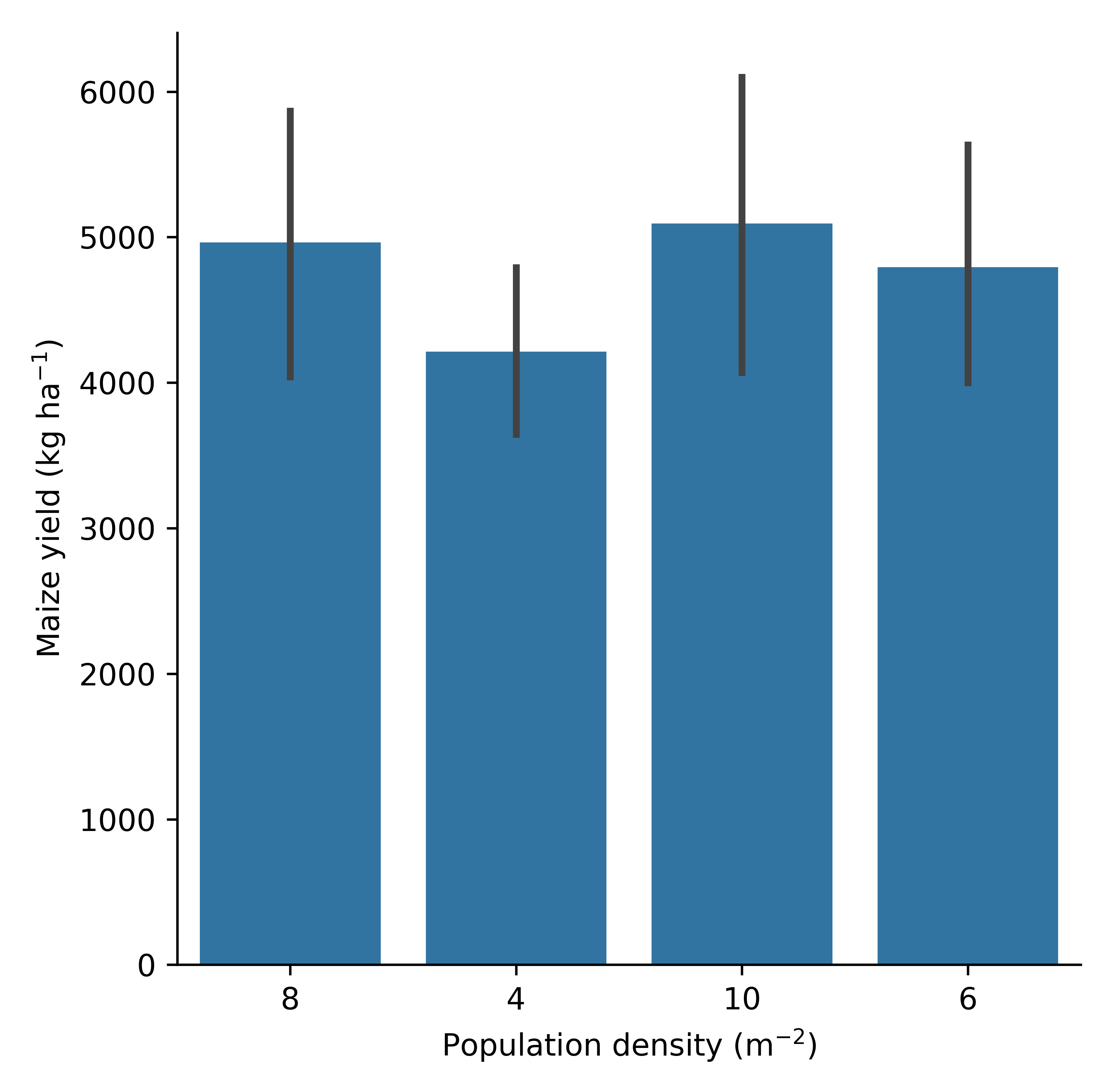
g= model.cat_plot(table = 'Report', y='Yield', x= 'Population', kind='bar')
g.set_axis_labels(r"Population density (m$^{-2}$)", r"Maize yield (kg ha$^{-1}$)")
g.set_titles("Yield vs population density")
Maize yield labeled plot (bar plot).
Passing a custom dataset
Because apsimNGpy plotting wraps Seaborn, you don’t need extra imports for most advanced visuals or after some advanced calculation on your dataset.
When you want to pre-compute values or reshape data yourself, use the table argument: it accepts None, a table name (str) from the model’s database, or a pandas.DataFrame.
table=None : uses self.results from the model.
table=”Report” : (str): loads that table from the DB.
table=df (DataFrame) : uses your custom, in-memory data.
The example below shows how to convert a numeric nitrogen Amount to an ordered categorical (for cleaner legends/ordering) and pass it straight into any plotter:
import pandas as pd
# Example: start from model results (or load a table)
df = model.results.copy() # or: df = model.get_simulated_output("Report")
# Convert nitrogen rate to an ordered categorical for consistent ordering/legend
bins = [0, 50, 100, 150, 200, 250]
labels = ["0–50", "51–100", "101–150", "151–200"]
# 1) Coerce to numeric
df = df.copy()
df["Nitrogen"] = pd.to_numeric(df["Nitrogen"], errors="coerce")
# 2) Handle NaNs (drop or label them)
# Option A: drop rows with invalid Amount
df = df.dropna(subset=["Nitrogen"])
# Option B: keep them and fill a placeholder after cut
# 3) Define consistent bins & labels
bins = [0, 50, 100, 150, 200] # strictly increasing
labels = ["0–50", "51–100", "101–150", "151–200"] # len=4 = len(bins)-1
# 4) Cut (older pandas? remove ordered= and set later)
df["N_rate_class"] = pd.cut(
df["Nitrogen"],
bins=bins,
labels=labels,
include_lowest=True,
right=True, )
# Optionally, sort by the new category
df = df.sort_values("N_rate_class")
# Use the custom DataFrame directly via `table=...`
# Example 1: line series
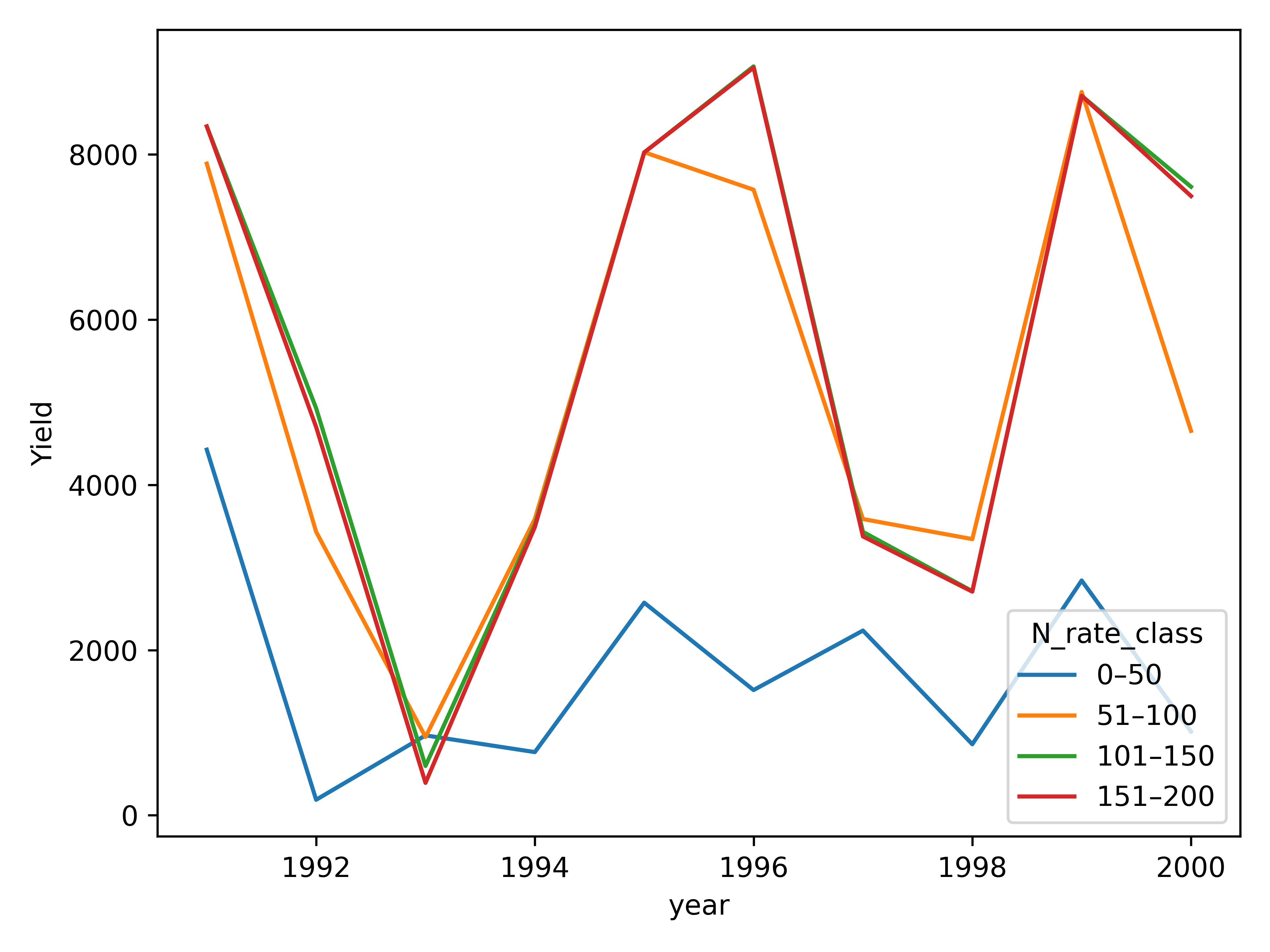
model.series_plot(
table=df,
x="year",
y="Yield",
hue="N_rate_class", errorbar=None
)
plt.tight_layout()
plt.savefig("series_ordered.png")
Maize yield ordered plot (Line plot)
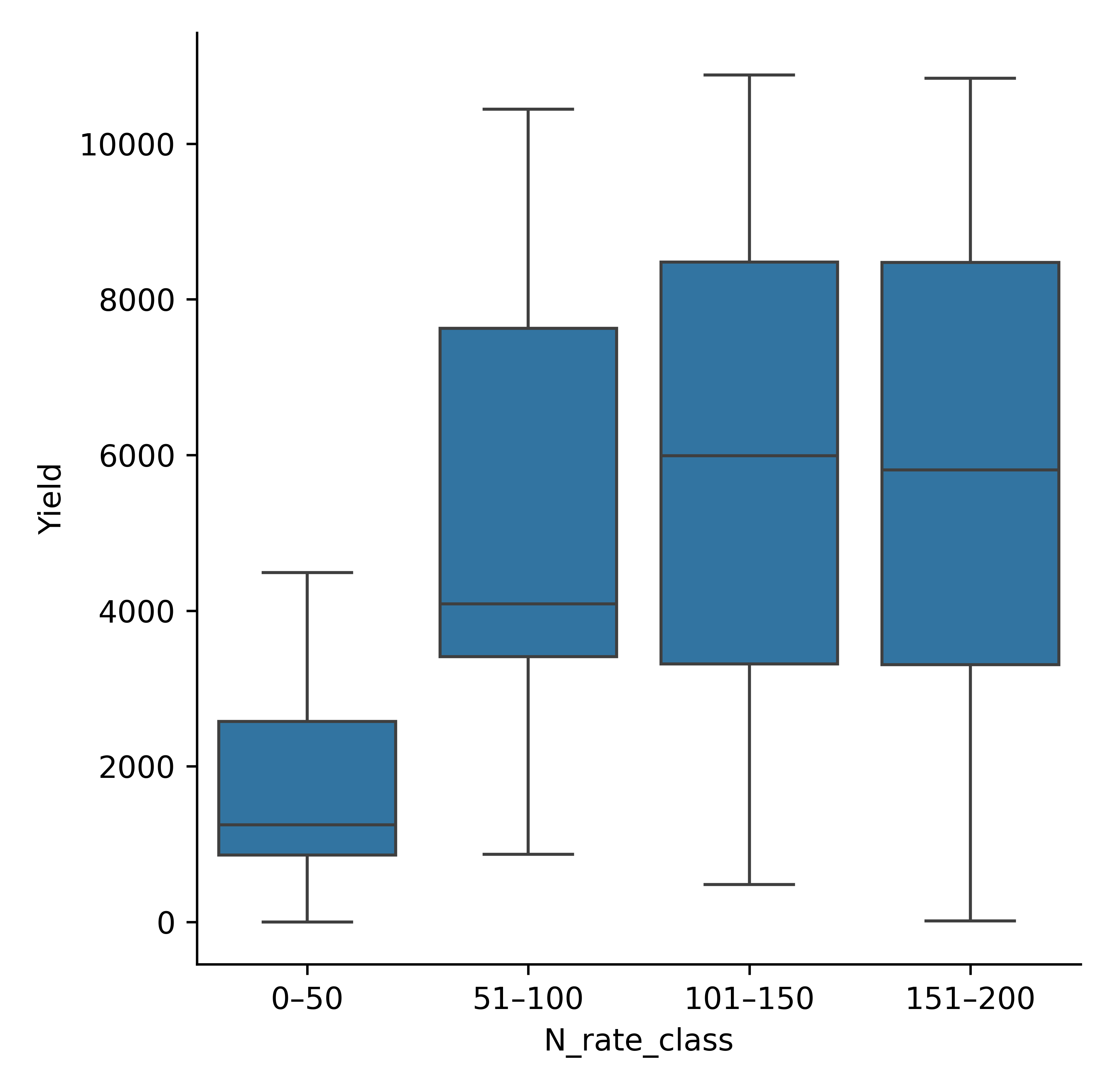
# Example 2: categorical plot (e.g., box/violin via catplot)
model.cat_plot(
table=df,
x="N_rate_class",
y="Yield",
kind="box"
)
plt.savefig(p/"binned_cat_plot.png", dpi =600)
Binned Maize yield (bar plot).
Meta info
Notebook generated by;
APSIM version: `APSIM 2025.8.7844.0`
apsimNGpy version 0.39.10.17
See also